3DGene: Gene Structure Database
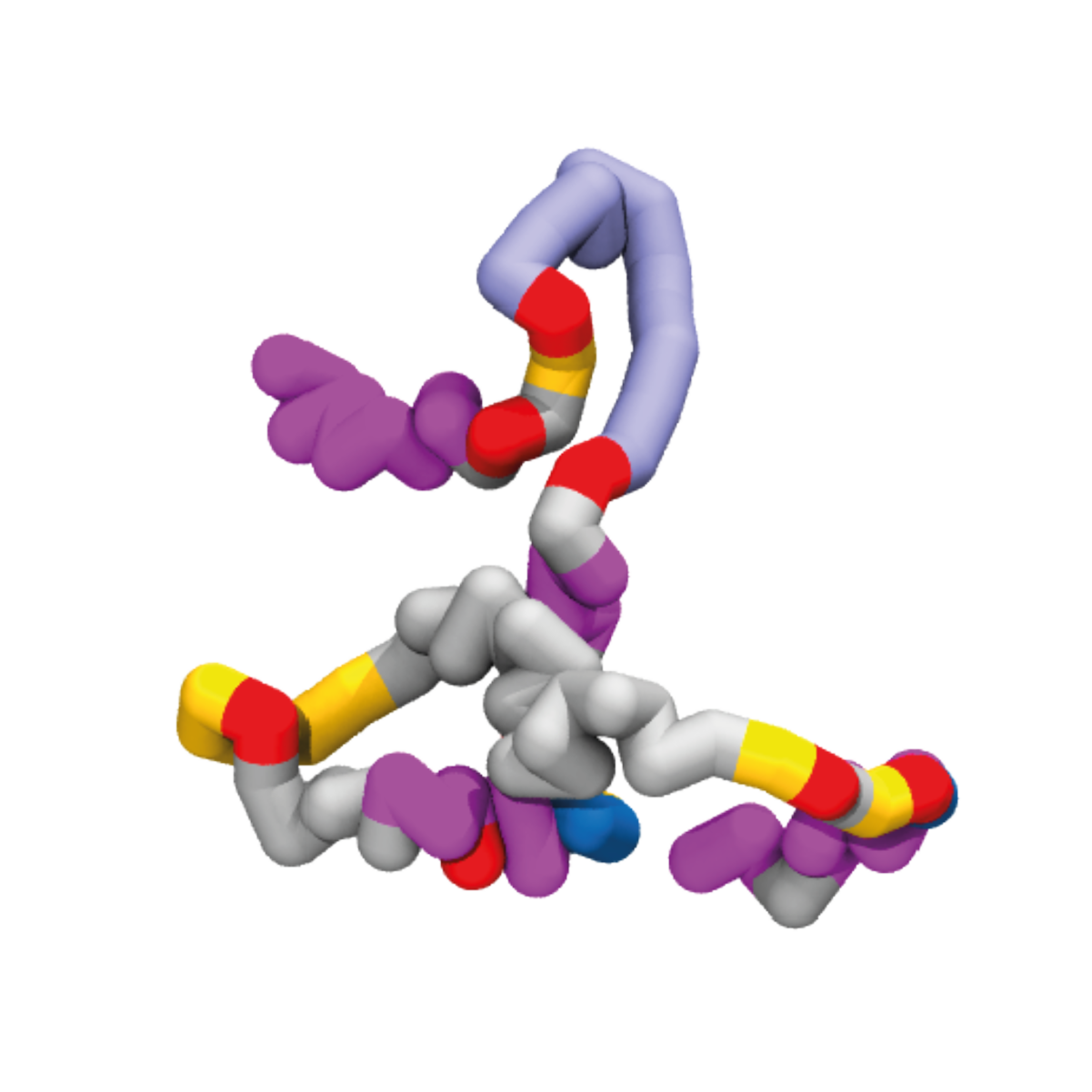
3DGene database provides a visualisation of the 3D conformation of 10,742 active genes in GM12878 human lymphoblastoid cells. The structure of each chromosome was simulated 600 times using the HiP-HoP framework; individual gene structures were extracted, and the three most common gene topologies (promoter-enhancer interaction networks) are available to view online with the average contact matrix. Simulation data for all genomic regions are available for download in the Downloads page.
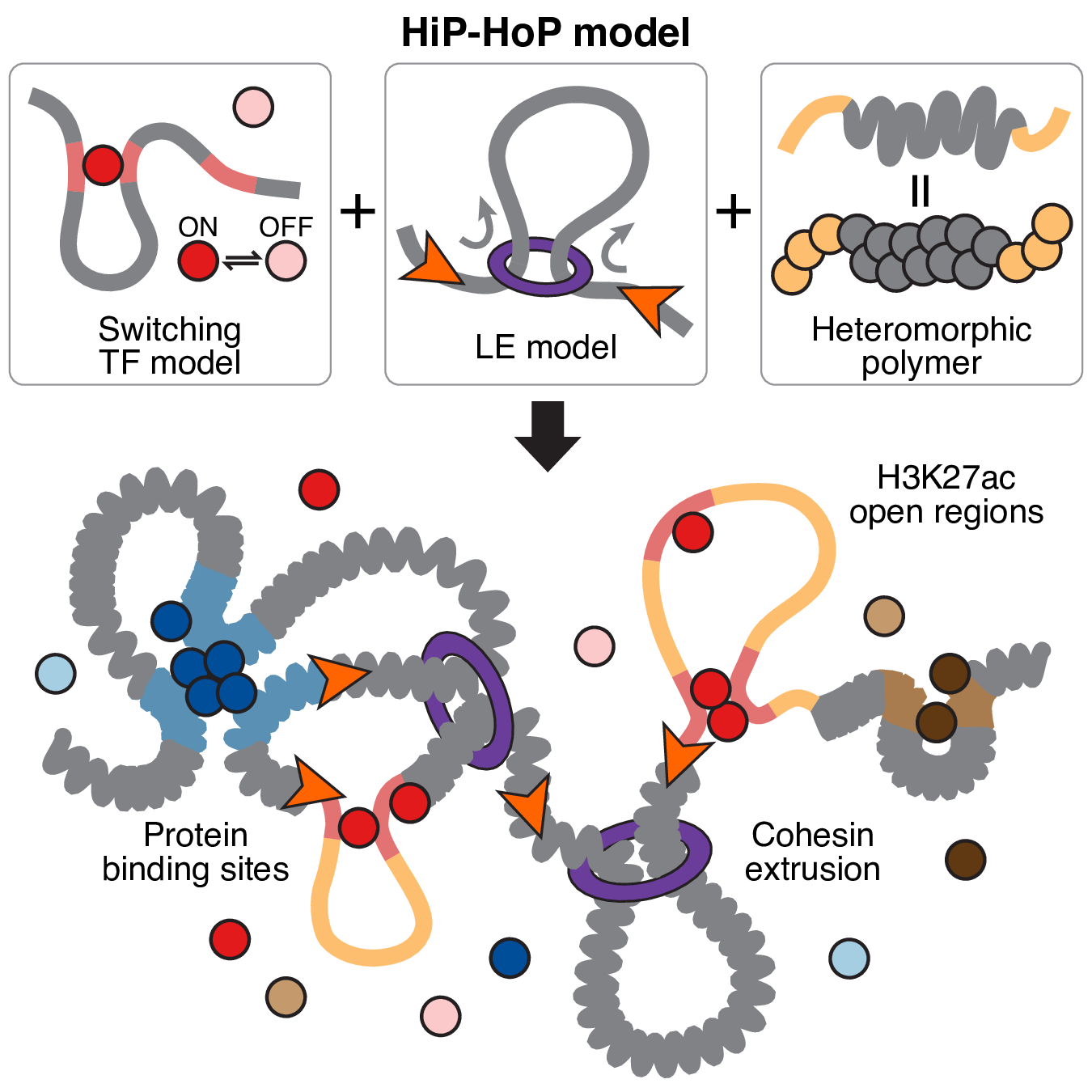
The HiP-HoP (Highly Predictive Heteromorphic Polymer) framework is a mechanistic polymer model that takes 1D epigenetic data (H3K27ac, H3K9me3, H3K27me3, ATAC, CTCF/cohesin binding sites) and predicts the 3D conformation of gene topologies. This model is built on three key foundations: switching transcription factor interactions, loop extrusion and a heteromorphic polymer.
Using this framework we predicted the 3D structure of the genome of GM12878 lymphoblastoid cells. Each chromosomal segment was simulated 600 times, providing insight into structural heterogeneity and enabling inferences to be made about the properties important for regulating chromatin folding.
An accompanying manuscript to 3DGene is available on Cell Genomics: Michael Chiang, Chris A. Brackley, Catherine Naughton, Ryu-Suke Nozawa, Cleis Battaglia, Davide Marenduzzo, Nick Gilbert. Genome-wide chromosome architecture prediction reveals biophysical principles underlying gene structure. Cell Genom. 2024 Dec 11;4(12):100698. doi: 10.1016/j.xgen.2024.100698. Epub 2024 Nov 25. PMID: 39591973.
- Brackley CA, Marenduzzo D, Gilbert N. Mechanistic modeling of chromatin folding to understand function. Nat Methods. 2020 Aug;17(8):767-775. doi: 10.1038/s41592-020-0852-6. Epub 2020 Jun 8. PMID: 32514111.
- Brackley CA, Gilbert N, Michieletto D, Papantonis A, Pereira MCF, Cook PR, Marenduzzo D. Complex small-world regulatory networks emerge from the 3D organisation of the human genome. Nat Commun. 2021 Oct 1;12(1):5756. doi: 10.1038/s41467-021-25875-y. PMID: 34599163; PMCID: PMC8486811.
- Chiang M, Brackley CA, Marenduzzo D, Gilbert N. Predicting genome organisation and function with mechanistic modelling. Trends Genet. 2022 Apr;38(4):364-378. doi: 10.1016/j.tig.2021.11.001. Epub 2021 Nov 29. PMID: 34857425.
- Chiang M, Forte G, Gilbert N, Marenduzzo D, Brackley CA. Predictive Polymer Models for 3D Chromosome Organization. Methods Mol Biol. 2022;2301:267-291. doi: 10.1007/978-1-0716-1390-0_14. PMID: 34415541.